My research interests focus on using novel computational techniques to enhance prediction of genotype-to-phenotype associations. My academic and research backgrounds are rooted in both biology and computer science, allowing me to integrate concepts and methods from both fields in my research. As an undergraduate at Brown University, I conducted research under Dr. Irina Arkhipova at the Marine Biological Laboratory (MBL). My project investigated the genome of a novel parasitic wasp whose body size is greatly reduced compared to related wasps. The findings from this study resulted in a co-authorship publication, and I presented these findings at the Mobile Genetic Elements conference hosted at the MBL. As a current graduate student at Duke University, I work with Dr. Allison Ashley-Koch on the genetic modifiers of sickle cell disease (SCD). Our lab is broadly interested in the genetic epidemiology of human genetic disorders, and my project and program will comprehensively prepare me for a career in academia. My project aims to impute multi-omics profiles in SCD patients, which will then enable prediction of renal outcomes in those patients, utilizing novel machine learning methods to strengthen the power of the imputation algorithms. My program and institution provide multiple opportunities for professional development, career exploration, skills development, and mentorship opportunities, in which I heavily engage. These opportunities also support my own personal goals of becoming a well-rounded scientist, and making science accessible to both the public and to future scientists. I am a first-generation college student: my family has always emphasized the value of education, and I want to help others in similar positions who want to pursue higher education. Overall, my goal is to become a scientist proficient in both research and mentorship, able to pose and investigate scientific inquiries and translate their results for the greater community to benefit.
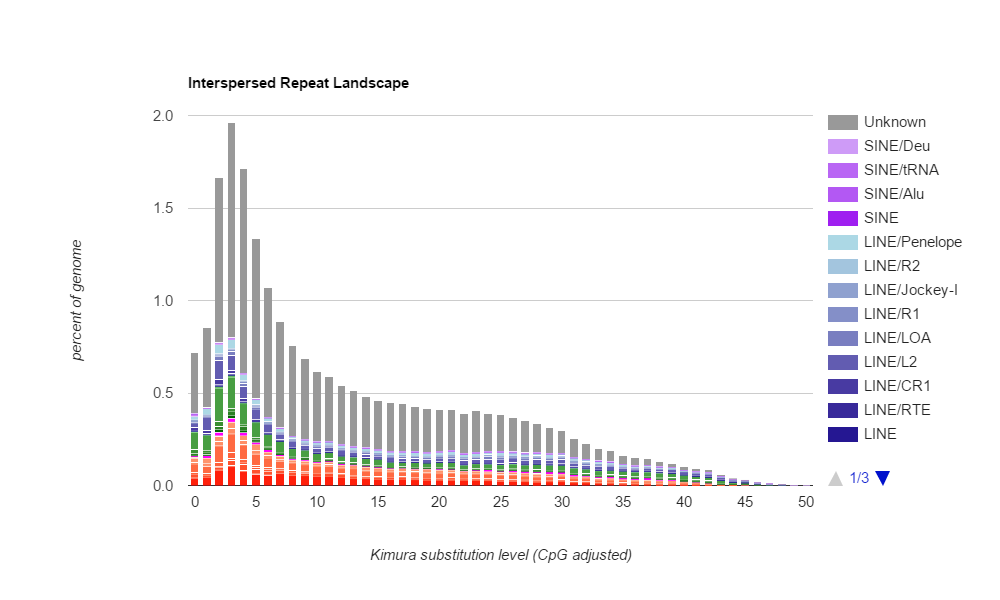
As an undergraduate student, I conducted research at the Marine Biological Laboratory over two summers. Under the guidance of Dr. Irina Arkhipova, I investigated the genome of a novel species of parasitic wasp (Megaphragma amalphitanum), focusing on how transposable elements (TEs) may have affected its evolution. Wasps in Megaphragma have microscopic body sizes, and our hypothesis was that TEs affected genome composition and contributed to body size reduction. I performed de novo detection and annotation of transposable elements in M. amalphitanum, writing and maintaining various programs to determine the wasp’s TE composition and history. Compared to related wasps, M. amalphitanum had similar TE composition but different TE activity, where it is currently experiencing less TE integration compared to its evolutionary past. This could be attributed to recently-acquired genome defense machinery that prevents rampant TE accumulation in its genome.
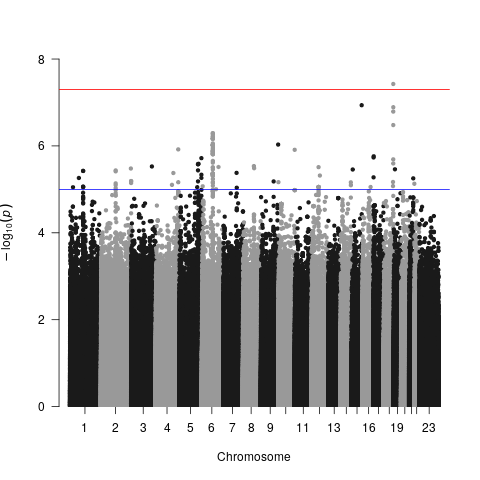
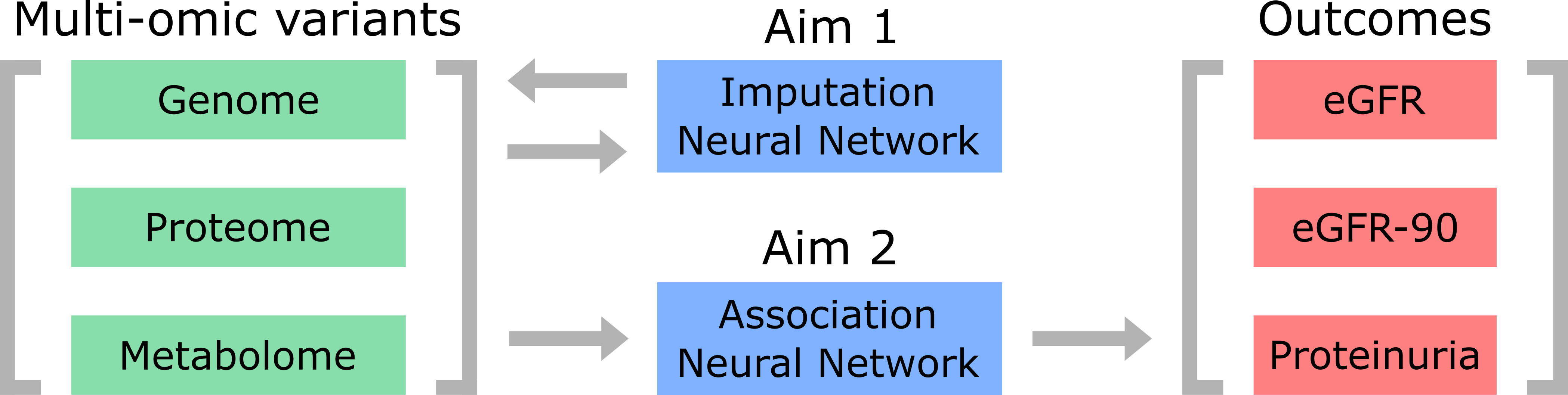
As a graduate student, I conduct research on the genetic modifiers of sickle cell disease (SCD). SCD pathogenesis varies widely between individual patients despite the constancy of a beta globin point mutation, suggesting that other factors influence SCD disease progression. In support of this research, our lab has collected data on SCD nephropathy (SCDN) and multi-omics profiles, which includes genomes, metabolomics, and proteomics. This data is organized in a well-structured SCD patient cohort as part of the NHLBI TOPMed program. In conjunction with several other TOPMed SCD cohorts, our aim is to better characterize omic factors contributing to SCDN progression. The omics data available for analysis are not always complete, and there exists potential gains from imputation of currently available data to boost the coverage and statistical power of these analyses. My project will develop deep learning methodologies to impute missing multi-omics data and predict SCDN outcomes given a patient’s existing set of multi-omics profiles. GWASes and meta-analyses of the TOPMed SCD cohorts have been performed, and the proposed work will utilize these results to construct, train, and test neural networks capable of both imputation and omics-phenotype association.
A select list of projects and publications are displayed below.